On June 6, Professor Dong Bo's team from Fang Zongxi Center for Marine Evo-Devo published a research paper titled Physics of Notochord Tube Expansion in Ascidian in PNAS. With a focus on the morphological development of the notochord tube in the ascidian at crucial evolutionary stages, the research reveals that the mechanical contraction force generated by the Cdc42 signaling-regulated cell cortical constriction ring, combined with the physical barrier created by intercellular tight junctions, plays a key role in the geometric shape and volume regulation of the lumen fluid within the cells.
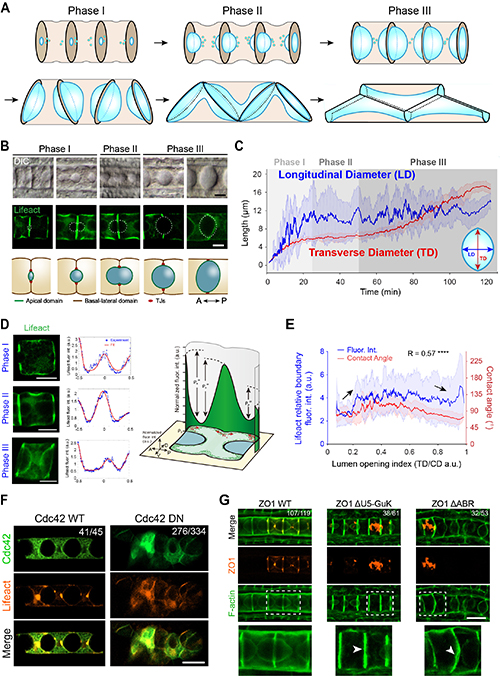
In this study, the researchers utilized an emerging tubulogenesis model in the ascidian notochord, in which lumen pockets first appeared between pairs of notochord cells and then coalesced into a multicellular tube (Figure 1A-B). This study first established a transgenic glass ascidian with notochord-specific expression of a fluorescent marker for the cell cortex (Lifeact-eGFP) to accurately quantify the geometric parameters of the lumen expansion process. By analyzing the distribution of notochord cortical fluorescence signals at different developmental stages, the researchers identified a specific structure of actin cortex contraction rings at the lumen edges. This structure demonstrates a clear correlation between the strength of the contraction rings and the geometric shape of the lumen. To figure out the upstream regulatory mechanisms of the cortical signals, the study systematically screened the Rho GTPase family proteins that are highly expressed in the notochord, and found that dominant negative mutations in Cdc42 resulted in severe deformities in notochord development and lumen formation. The researchers then focused on the mechanism by which cells regulate the lumen volume; through the expression of domain deletion mutants of the tight junction protein ZO1, they discovered a significant reduction in lumen volume.
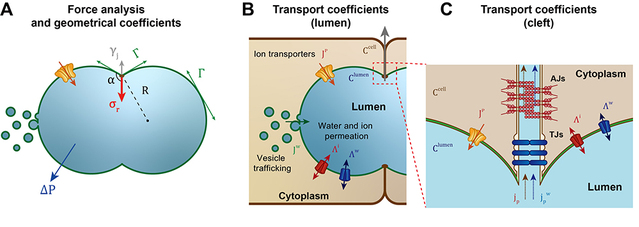
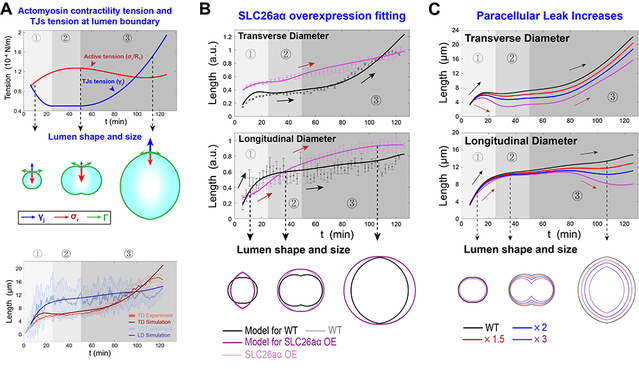
Based on the cellular regulatory mechanisms for lumen dynamics identified in this study, previous research on the mechanisms of lumen formation and the conservation law, the researchers established a general physical model for lumen expansion under the synergistic action of cells and lumen fluid. They then performed numerical solutions for the established model. Using geometric data of the lumen and contraction ring intensity variations obtained from experimental observations, they simulated the dynamics of wild-type lumen expansion. Building on this finding, the study successfully predicted and explained the origins of the dynamic phenotypes resulting from disruptions during the lumen formation process. Furthermore, the team predicted the post-interference luminal dynamic phenotypes under various scenarios, identified the specific regulatory mechanisms of the complex cellular regulatory network over the lumen, and further provided theoretical thresholds for the biological processes involved.
In summary, this study elucidated the key cellular regulatory mechanisms underlying lumen expansion and homeostasis maintenance, thereby laying an important foundation for systematically explaining the biological regulatory network of biological lumens. The bile canaliculi in the human liver exhibit a structure remarkably similar to that of the notochord lumen in ascidians. The dynamic model of lumen expansion established in this study could provide a theoretical basis for identifying and treating the pathogenesis of bile canaliculus-related diseases.