Recently, Professor Li Qi and Professor Liu Shikai from the Laboratory of Shellfish Genetics and Breeding at Fisheries College in collaboration with researchers from the University of Edinburgh, have made new progress in the research field of genome diversity in bivalves. Their findings have been published online in Cell Reports titled “Haplotype-resolved genomes provide insights into the origins and functional significance of genome diversity in bivalves.”
Bivalves, such as oysters, scallops, and mussels, are crucial species in China’s marine aquaculture industry. Their genomes exhibit exceptional genetic diversity, which surpasses that of many other organisms. Understanding the origins and functions of the genetic diversity is vital to unraveling the relationship between genetic innovation and adaptive evolution. Additionally, exploring the mechanism behind this genome diversity is essential for overcoming current bottlenecks in genome-assisted breeding technologies and developing breakthrough aquaculture varieties.
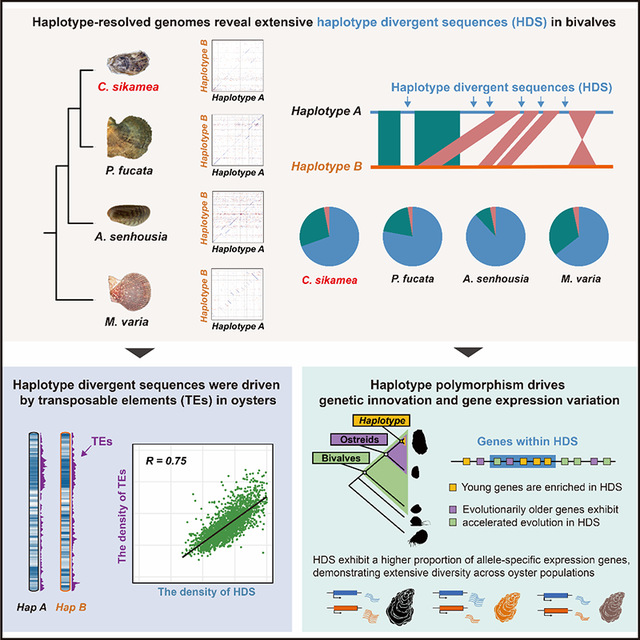
The research team constructed the first haplotype-resolved genome for Crassostrea sikamea. Through comparison of haplotype-resolved genomes across oysters, scallops, mussels, and pearl oysters, the team discovered the widespread existence of haplotype divergent sequences (HDSs) in bivalves. Based on this discovery, the team proposed the hypothesis that bivalves maintain a unique “gene pool” by preserving extensive individual-specific genome diversity, which enables adaptive evolution in the highly variable marine environment.
The study revealed that HDSs in bivalves serve dual functions: first, as “incubators” for new genes, bivalves can drive the generation of innovative genetic elements; second, by accelerating the evolutionary rate of conserved genes, they can facilitate the adaptive evolution of key physiological functions, including osmotic regulation and immune defense. This adaptive evolutionary strategy enables bivalves to maintain the stability of basic life processes while swiftly responding to environmental pressures. Furthermore, the study provided new insights into the relationship between genome diversity, phenotypic plasticity, and adaptive evolution by evaluating the evolutionary conservation of whole-genome sequences and conducting transcriptional dynamics analysis of cross-group spatiotemporal interactions.