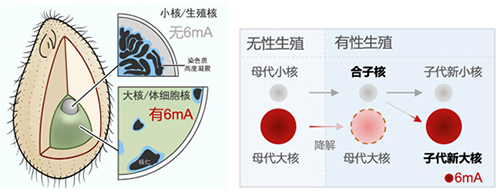
In May 2025, the protozoology team led by Professor Gao Shan from Marine Biodiversity and Evolution Research Institute published their latest research titled Identification and characterization of the de novo methyltransferases for eukaryotic N6-methyladenine (6mA) in the journal Science Advances. With the single-celled eukaryote Tetrahymena thermophila as the material, the researchers utilized a unique time window where the zygotic nucleus without 6mA developed into a new macronucleus with 6mA during its conjugative reproduction process. The study identified the MT-A70 family proteins AMT2 and AMT5 as de novo methyltransferases for 6mA, and further revealed their critical roles in the establishment of 6mA and normal cellular development.
The study found that AMT2 and AMT5 are significantly expressed during the later stages of sexual reproduction and are specifically distributed in the new large nucleus, closely aligning with the timing and location of de novo methylation of 6mA (Figure 2A-B). To verify whether these two genes possess de novo methyltransferase activity, the research team constructed homozygous lines for single or double knockout of AMT2 and AMT5. Immunofluorescence staining and small molecule DNA mass spectrometry results both indicated that the level of 6mA in the knockout lines was significantly decreased compared to the wild type, by approximately 80% (Figure 2C). Complementation experiments demonstrated that the wild-type gene complement could effectively restore 6mA levels, while complementation with mutations at key active sites of the enzyme could not restore the levels. This indicated that the decrease in 6mA caused by the knockout is directly related to the loss of methyltransferase activity (Figure 2C).
To further analyze the changes in the 6mA patterns in the knockout lines, the team conducted single-molecule real-time sequencing (SMRT-CCS) on the progeny new large nuclei obtained through flow sorting (Figure 2D). This approach demonstrated that AMT2 and AMT5 are crucial for the correct establishment of the 6mA profile in the new large nucleus.

Further analysis revealed that during the development of the zygotic nucleus into the progeny new large nucleus, AMT1 can partially compensate for the functions of AMT2 and AMT5 in the absence of these two genes, thereby contributing to de novo methylation. However, due to the lack of a foundational preparation by the de novo methyltransferases, the number of catalytic sites available for AMT1 is limited, and its methylation efficiency is low. This further highlighted that the catalytic activity of AMT2 and AMT5 plays a crucial role and serves as the rate-limiting step for the de novo establishment 6mA in the new large nucleus.
Based on the above results, the team proposed a regulatory pathway model for 6mA (Figure 3). In wild-type Tetrahymena, during the development of the zygotic nucleus, which is devoid of 6mA, into a new large nucleus that contains 6mA, the de novo methyltransferases AMT2 and AMT5 catalyze the conversion of unmethylated adenine to hemi-methylated adenine. Subsequently, the maintenance methyltransferase AMT1 further converts it into fully methylated adenine. After entering asexual reproduction, the sustained high expression of AMT1 ensures stable inheritance of 6mA during cell division.
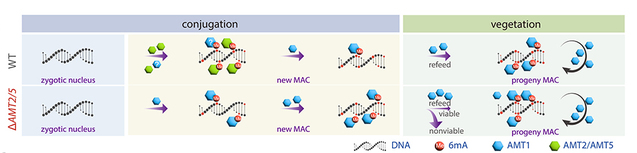
In cells lacking AMT2 and AMT5, AMT1 exhibits a (weaker) activity as both a de novo methyltransferase and a maintenance methyltransferase, capable of adding 6mA at only a few loci, resulting in significantly lower methylation levels. Once the cells enter asexual reproduction, the increase in AMT1 levels allows for the methylation of a large number of unmethylated loci; however, there may still be disruptions in the methylation patterns.
This study for the first time identified and characterized de novo methyltransferases that mediate the addition of 6mA in eukaryotes, revealing that the 6mA modification follows a two-step methylation pattern of establishment and maintenance. This finding forms an interesting correspondence with classical 5-methylcytosine (5mC): both modifications recognize palindromic sequences (ApT for 6mA and CpG for 5mC), and are inherited in a semi-conservative manner; and they rely on a two-step methylation process to complete the modification. Considering that 6mA is primarily found in unicellular eukaryotes while 5mC is widely distributed across multicellular organisms, this study offers a new perspective for understanding the differentiation and evolution of eukaryotes from the perspective of DNA methylation.







