Recently, the research group led by Professors Su Ying and Zhao Long from the Institute of Evolution and Marine Biodiversity, in collaboration with BGI-Qingdao, has achieved significant progress in the field of zebrafish heart regeneration. Their findings have been published in Nature Communications on April 19th under the title An Organ-wide Spatiotemporal Transcriptomic and Cellular Atlas of the Regenerating Zebrafish Heart.
Zebrafish possess a remarkable capacity for heart regeneration. Following injury, surviving cardiomyocytes near the wound undergo cellular biological events such as dedifferentiation, proliferation, and redifferentiation to generate new cardiomyocytes, ultimately achieving the complete restoration of both the structure and function of the damaged heart. Elucidating the cellular dynamics and molecular regulatory networks involved in this process is regarded as a crucial breakthrough for tackling the challenges in cardiac regenerative medicine.
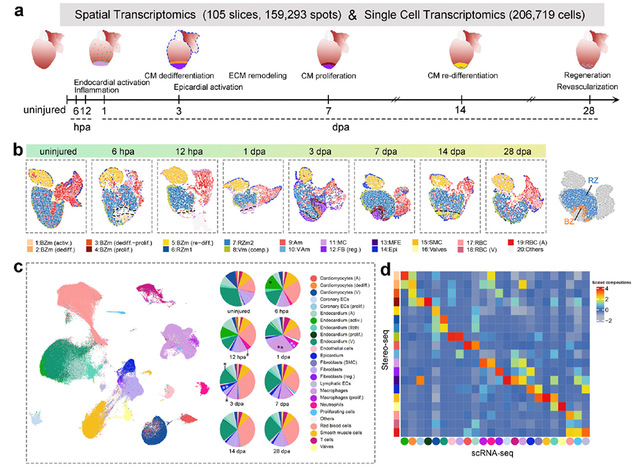
This study employed spatial transcriptomics (Stereo-seq) and single-cell transcriptomics (scRNA-seq) technologies to perform whole-organ dynamic analysis of the zebrafish heart across 8 key time points before and after injury, thereby creating a single-cell and spatiotemporal transcriptomic atlas of zebrafish heart regeneration. This atlas systematically captured and described the spatiotemporal interaction networks among various key cardiac cell types, including cardiomyocytes, endothelial cells, immune cells, and fibroblasts. Furthermore, combined with molecular and cellular experiments, the study specifically investigated the dynamic expression and functional necessity of the tpm4a gene during the redifferentiation event of newly formed cardiomyocytes, revealing the importance of the gene in heart regeneration.
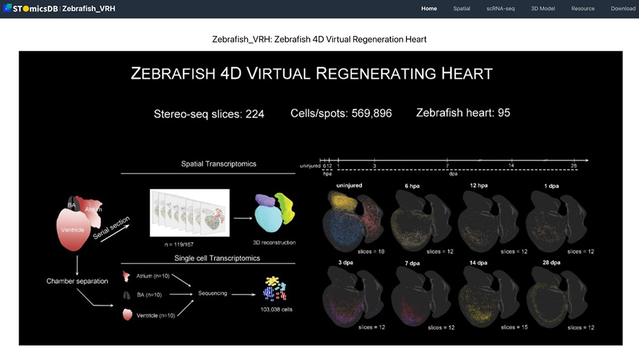
Given the significant differences in cardiac regenerative capacity across various species, this study identified evolutionarily conserved genes, such as ifrd1 and atp6ap2, which are associated with the functional divergence between regenerative and non-regenerative hearts in different species. These findings provide potential targets for research into clinical therapies for cardiovascular diseases. Finally, the research team utilized the omics data to construct a 3D transcriptomic atlas of the zebrafish heart and a 4D dynamic model of regeneration, which intuitively visualized the spatiotemporal evolution patterns of cell types and gene expression during the heart regeneration process. This model has been made globally accessible through the interactive database Zebrafish_VRH (https://db.cngb.org/stomics/zebrafish_VRH/), offering powerful tools and resource support for regenerative medicine research.