DNA N6-methyladenine (6mA) has been reported to participate in critical biological processes such as transcriptional regulation in eukaryotes [1-6]. However, the relationship between 6mA and transcription exhibits significant variations across different eukaryotic organisms, and the specific mechanisms by which 6mA engages in transcriptional regulation remain incompletely elucidated. Tetrahymena thermophila, an important single-celled eukaryotic model organism, has been proved to possess high 6mA levels catalyzed by the methyltransferase AMT1 (Adenine Methyltransferase 1). The positive correlation between 6mA and transcription makes this organism an ideal system for studying 6mA-mediated transcriptional regulation mechanisms.
In January 2025, the research team led by Professor Gao Shan from the Institute of Marine Biodiversity and Evolution at OUC published a study titled Methyl-dependent auto-regulation of the DNA N6-adenine methyltransferase AMT1 in the unicellular eukaryote Tetrahymena thermophila in Nucleic Acids Research. Utilizing Tetrahymena as a model system, this research revealed the auto-regulatory and transcriptional control mechanisms of the 6mA methyltransferase AMT1, investigated its dynamic distribution and physiological functions during conjugation, and offered critical insights into 6mA as an epigenetic mark participating in transcriptional regulation and other physiological processes in eukaryotes. These findings establish a theoretical foundation for future study of 6mA's functional regulatory mechanisms in eukaryotic systems.
In previous work, the team identified AMT1 as the primary 6mA methyltransferase in Tetrahymena through studies using this organism as a model system. It was found that deletion of the AMT1 gene would result in slowed cell growth and abnormally enlarged contractile vacuoles. However, the molecular mechanisms by which AMT1 regulates gene expression to influence cellular growth and development remained unknown.
This study revealed that mutation of critical catalytic sites in AMT1 not only caused a significant genome-wide reduction in 6mA levels (Figures 1A-C) but also notably decreased the mRNA and protein expression of the AMT1 gene itself (Figures 1D-E). Analysis of the chromatin environment surrounding the AMT1 gene identified characteristic 6mA-associated features, including stable nucleosome occupancy, elevated levels of H3K4me3 and H2A.Z (Figure 1F), and relatively high 6mA enrichment (Figure 1G). Based on these findings, researchers hypothesize that the transcriptional activity of the AMT1 gene may be regulated by its own 6mA modifications.
The team further conducted single-base resolution SMRT-CCS sequencing on AMT1 mutant strains. The results demonstrated significant reductions in 6mA levels post-AMT1 mutation (Figures 2A-B), in line with previous 6mA immunofluorescence staining and mass spectrometry data. Notably, 6mA levels on the AMT1 gene itself showed significant declines (Figures 2D-F). Transcriptome sequencing revealed corresponding declines in AMT1 mRNA expression (Figure 2D). These findings suggest that AMT1 regulates its own gene's 6mA levels through catalytic activity, subsequently influencing transcriptional output. Rescue experiments with ectopic expression of wild-type AMT1 protein confirmed that complementation restored both 6mA levels and mRNA expression of the AMT1 gene (Figure 2G), demonstrating a 6mA-mediated positive feedback loop for transcriptional activation. This research has provided additional evidence for 6mA's role in transcriptional regulation, demonstrating its potential as an epigenetic mark for transcriptional activation in cellular physiological processes (Figure 3).

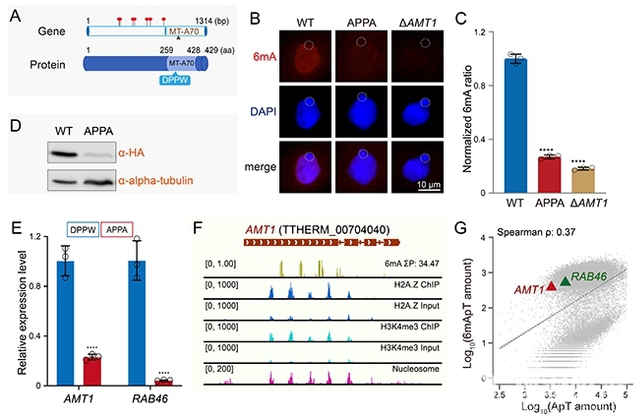
Figure 1
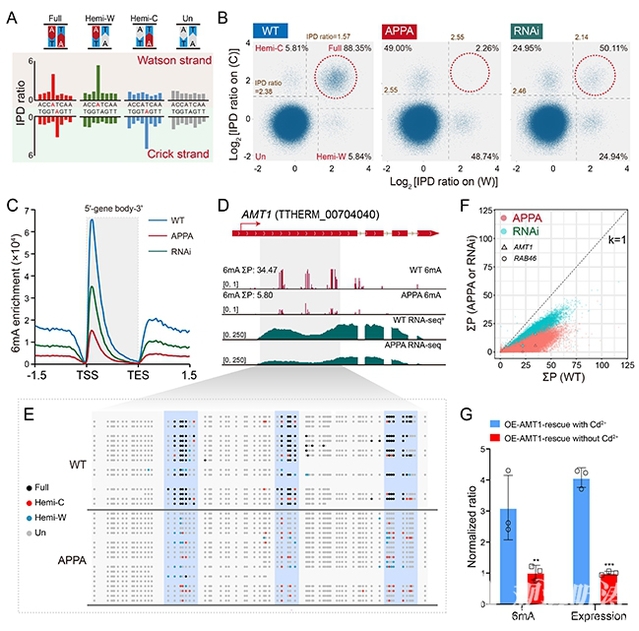
Figure 2
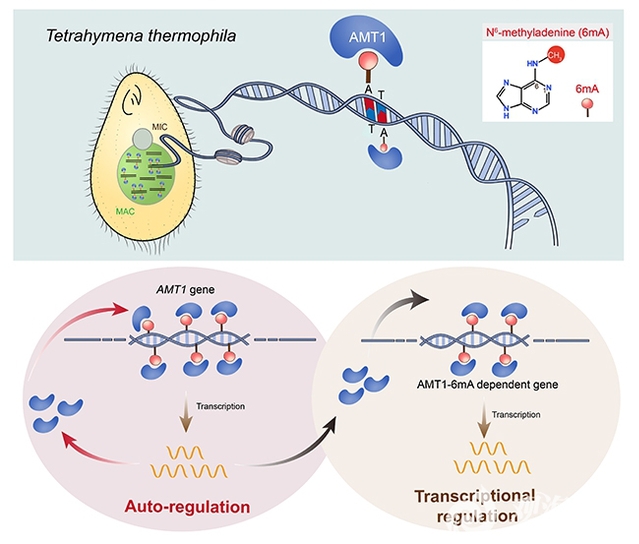
Figure 3







