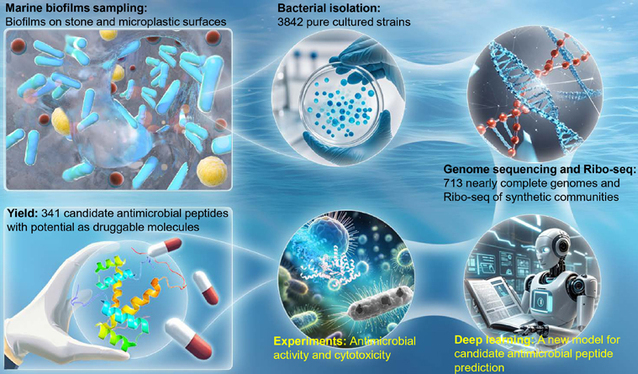
Recently, Professor Zhang Weipeng from the Institute of Marine Biodiversity and Evolution, and Associate Professor Ding Wei from the College of Marine Life Sciences published a research paper titled Bioprospecting of culturable marine biofilm bacteria for novel antimicrobial peptides in the journal iMeta. By using new methods, they explored the potential of marine biofilm bacteria to produce antimicrobial peptides, expanded the scope of antimicrobial peptide compounds, and provided a new perspective for the research and development of antibacterial drug-derived molecules.
Antimicrobial peptides are amphiphilic small molecule polypeptides produced by animals, plants and microorganisms, which have broad-spectrum antibacterial effects. In the marine environment, biofilms are microbial communities attached to any submerged substrates, such as artificial substrates, stone surfaces, microplastics or animal viscera. Due to their unique ecological niches and metabolic characteristics, biofilm bacteria hold great potential for the discovery of novel antimicrobial peptides. This research focuses on bioprospecting culturable marine biofilm bacteria for novel antimicrobial peptides.
In this study, bacterial strains were isolated from marine biofilms and their genomes were sequenced, thus constructing a library of culturable marine biofilm bacteria. A novel method combining ribosome sequencing (Ribo-seq) with a deep learning model was applied to predict antimicrobial peptides. The introduction of Ribo-seq analysis enhanced the filtering process for sequencing, enabling more accurate identification of small open reading frames (sORFs) prior to the prediction of antimicrobial peptides. Then, a deep learning model consisting of a convolutional neural network, a bidirectional long short-term memory layer and an attention layer was created. Among 80,430 expressed sORFs, 341 were identified as candidate antimicrobial peptides. Among the 60 chemically synthesized candidate antimicrobial peptides, 90% (54) showed antibacterial activity and strong inhibitory effects on a variety of drug-resistant human pathogenic bacteria. Cytotoxicity and hemolytic tests on 12 of these highly potent antimicrobial peptides demonstrated relatively low toxicity. Finally, the structures of the antimicrobial peptides were explored through AlphaFold2 prediction and circular dichroism spectroscopy, and their action targets were also observed. The bactericidal mechanisms of four highly potent candidate antimicrobial peptides targeting the cell membranes of Staphylococcus aureus were identified.