The research team led by Professor Li Dehai from the School of Medicine and Pharmacy and the Key Laboratory of Marine Drugs, in collaboration with the team led by Professor Zhang Yuzhong from the College of Marine Life Science and the Key Laboratory of Marine Biodiversity and Evolution, has published their latest research outcome in Nature Communications. The research paper is titled Characterization and structural analysis of a versatile aromatic prenyltransferase for imidazole-containing diketopiperazines.
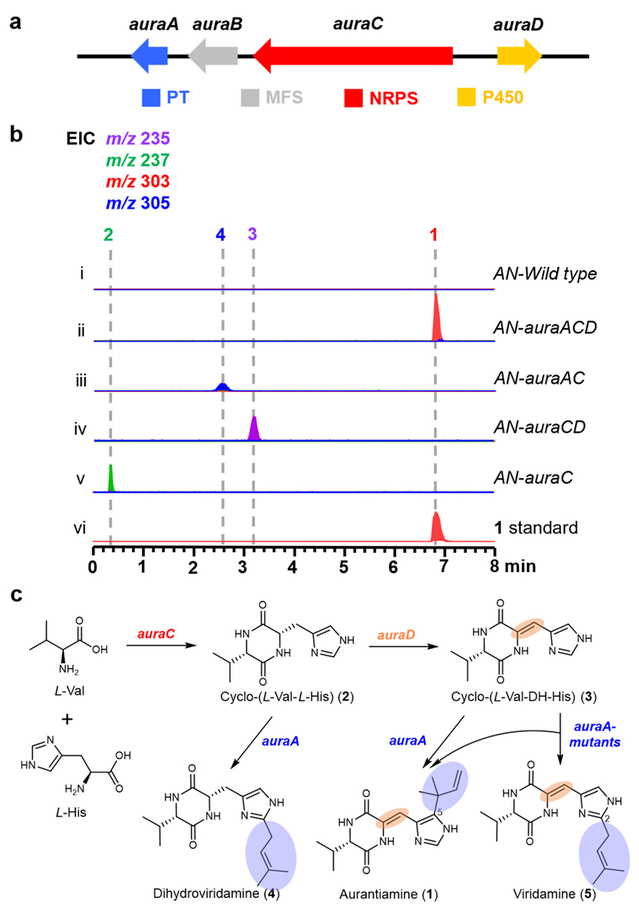
Researchers activated silent genes in the marine-derived fungus Penicillium solitum HDN11-131 and obtained the DKP compound aurantiamine, which features imidazole-C5-trans-dimethylallylation. To further study the process of imidazole prenylation, the biosynthetic gene cluster aura of aurantiamine was first identified in P. solitum HDN11-131. This gene cluster includes genes encoding DMATS family PT (AuraA), MFS transporter (AuraB), bimodular NRPS (AuraC), and P450 monooxygenase (AuraD). Through systematic heterologous expression in Aspergillus nidulans, the biosynthetic pathway of aurantiamine was identified. In the heterologous strain AN-auraAC, which contains the auraA and auraC genes, dihydroveridamine, a new substitution pattern product catalyzed by AuraA with imidazole-C2-cis-prenylation, was discovered. In vitro enzymatic experiments confirmed that AuraA can catalyze the dehydrogenation of the cyclic dipeptide cyclo-(L-Val-DH-His) to produce the imidazole-C5-trans-prenylated product 1, as well as catalyze the cyclic dipeptide cyclo-(L-Val-L-His) to generate the imidazole-C2-cis-prenylated product 4.
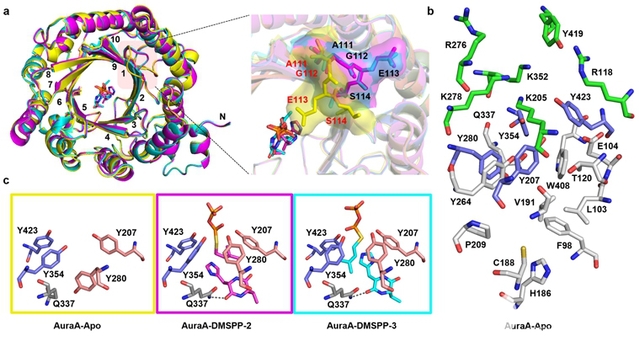
Researchers studied the structure of AuraA and its two ternary complexes (AuraA-DMSPP-2 and AuraA-DMSPP-3), systematically analyzing the conformational changes of residues within the catalytic pocket and the substrate binding modes. The research further revealed the influence of the relative positions and distances between the prenyl donor and acceptor on different catalytic modes and products, elucidating the structural basis and molecular mechanism of catalytic selectivity for AuraA-mediated imidazole prenylation. This study provides a new enzymatic tool for the alkylation modification of imidazole-based small-molecule drugs, and lays a structural foundation for understanding the recognition and catalytic mechanisms of the DMATS family of enzymes and improving their engineering.