Recently, the research team led by Professor Wang Min from the College of Marine Life Sciences at Ocean University of China (OUC) published their latest findings in Advanced Science. Their article, “Diversity and Ecological Potentials of Marine Viruses Inhabiting Continental Shelf Seas”, focuses on planktonic DNA viruses in the eastern continental shelf seas of China (ECSSC). Over six years, this virome-based study systematically reveals the diversity and spatiotemporal dynamics of these viral communities and their potential impacts on biogeochemical cycles. The study also establishes the world’s first genome dataset of DNA viruses from continental shelf seas, providing an important complement to existing global datasets of marine planktonic DNA viruses.
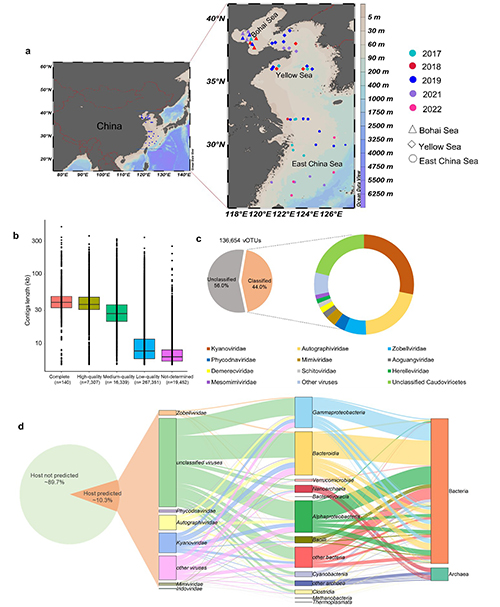
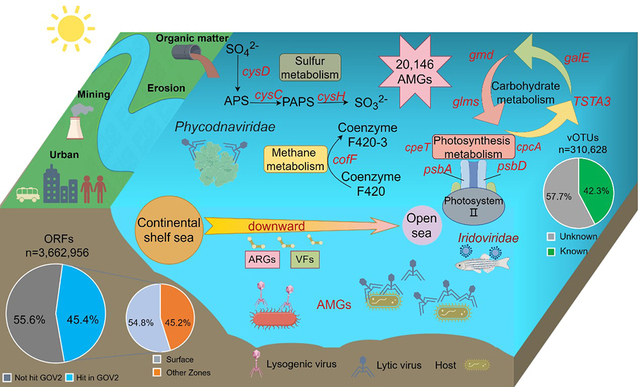
In this study, researchers found viruses from the ECSSC exhibit notable novelty: about 77% of the viral clusters are unique to the region, indicating that the ECSSC harbors a large number of previously unrecognized viral lineages. The study to the planktonic DNA virome over six years further reveals pronounced spatiotemporal patterns in DNA viral community structure: viral diversity in the Bohai Sea and Yellow Sea is higher than in the East China Sea, and viral diversity in summer is markedly higher than in winter; the major pathogens of fish and crustaceans, Iridovirus, were detected in the Bohai Sea and the Yellow Sea. In total, more than 20,000 virus-encoded auxiliary metabolic genes (AMGs) were identified, mainly enriched in pathways related to carbohydrate, sulfur metabolism, photosynthesis, and methane metabolism. AMGs related to polysaccharide synthesis and degradation, such as galE andgmd, and AMGs involved in sulfur metabolism, such as cysH and scyD, are significantly more abundant in the ECSSC than in open-ocean, polar, and hadal environments. This suggests that viruses in continental shelf seas may contribute more strongly to these elemental cycles. In addition, genes related to photosynthesis, such as psbA and cpeT, are also highly enriched in the ECSSC, further indicating that viruses may modulate carbon fixation in their autotrophic hosts. A total of 220 antibiotic resistance genes and 207 virulence factors were identified, with genes conferring resistance to vancomycin and polymyxins being particularly abundant. These genes show a clear pattern of “nearshore enrichment with offshore decline”, which is strongly associated with the intensity of human activities. This work provides important new data for understanding the diversity and ecological functions of viruses in continental shelf seas and lays a foundation for subsequent studies on virus-driven biogeochemical cycling in these regions.
Additionally, Professor Wang Min’s team also published their findings in Science China: Earth Sciences in an article entitled “Diversity, distribution and ecological potentials of Oceanospirillales-associated viruses in the global ocean”.
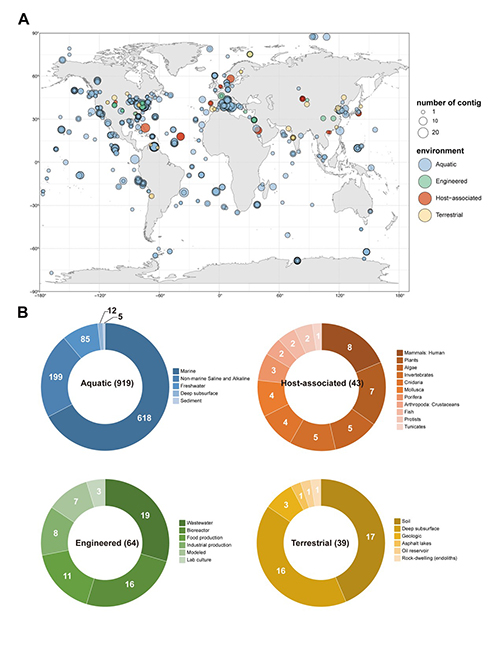
Species of the bacterial Order Oceanospirillales are widely distributed in polar and hadal environments. They exhibit the ability to metabolize alkane and play a crucial role in the bioremediation of hydrocarbon pollution. However, current knowledge of their viruses has largely been limited to the 7 isolated phages, lacking systematic studies on Oceanospirillales-associated viruses. In this study, the authors carried out the first global, systematic investigation of the genomic diversity, distribution, and ecological potential of uncultivated Oceanospirillales-associated viruses. They identified 3,273 uncultivated vital contigs linked to Oceanospirillales, including 783 high-quality genomes. These viruses exhibit the highest abundance in temperate and tropical marine epipelagic zones, as well as in polar regions. A total of 54 genes related to alkane metabolism were identified from these viral genomes, suggesting that Oceanospirillales-associated viruses may play important potential roles in the marine alkane cycle.








