Recently, Professor Yu Rilei from the School of Medicine and Pharmacy at Ocean University of China (OUC) , together with Professor Zhao Yan from the Institute of Biophysics, Chinese Academy of Sciences (CAS), published an article entitled “Deep Learning-Driven Discovery and Mechanism of Action Study of a Minimalist Conopeptide Targeting α7 Nicotinic Acetylcholine Receptor” in Acta Pharmaceutica Sinica B.
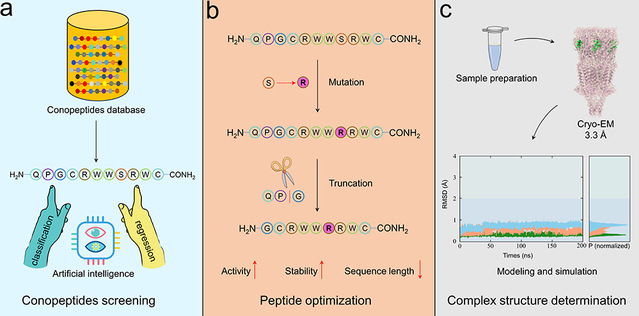
Marine cone snails produce a diverse array of bioactive conopeptides, comprising both disulfide-rich conotoxins and disulfide-poor peptides, which represent valuable resources for drug development and the design of neurochemical tools. While modern venomics approaches have dramatically expanded conopeptide discovery, their corresponding pharmacological characterization has lagged behind. Traditionally, the identification of functionally specific conopeptides still largely relies on random screening through labor-intensive electrophysiological experiments. In recent years, the introduction of high-throughput screening methods, including comprehensive venomics and phage display technologies, has significantly advanced the pharmacological development of bioactive peptides. Nevertheless, there remains an urgent need for rapid, high-throughput, and function-oriented screening strategies, which are of great scientific and practical value for efficiently elucidating the functions of conopeptides and identifying promising conopeptides.
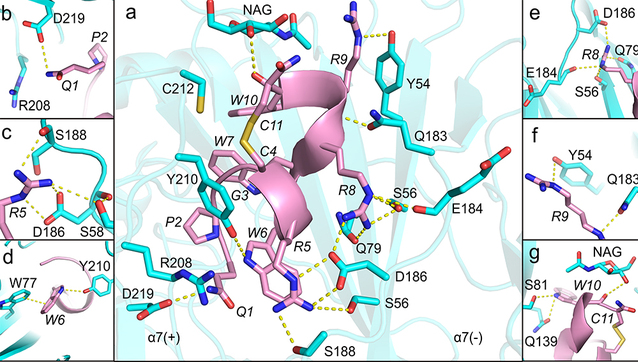
In this study, the research team developed an integrated pipeline combining deep learning, structural biology, computational modeling, and medicinal chemistry, successfully accelerating the discovery and optimization of conopeptides targeting the α7 nAChR, a key ion channel receptor closely associated with a variety of neurodegenerative diseases. Based on the ESM-2 protein language framework, they developed a deep learning-based screening model, enabling efficient screening of 689 disulfide-poor conopeptides and identified an active candidate molecule, SS1. Subsequent structure–activity relationship studies optimized the framework and yielded [S8R]SS1, a minimalist peptide with nanomolar potency that exhibits enhanced selectivity and improved stability. To gain mechanistic insights, the team employed cryo-electron microscopy (cryo-EM) to resolve the three-dimensional structure of the α7 nAChR and [S8R]SS1 complex, revealing its unique binding mode and mechanism of action through molecular dynamics simulations. This work not only provides a new methodological framework for the development of α7 nAChR-targeting marine peptides but also offers a more efficient route for drug discovery. Most importantly, it demonstrates how deep learning can be leveraged to achieve function-oriented screening of natural peptides under data scarcity, and how computational modeling and molecular simulations can effectively complement cryo-EM in resolving highly flexible peptide-receptor complexes, thereby significantly accelerating the development of natural peptide therapeutics.








